Platform
Platform
Platform to run Bayesian Networks and Machine Learning Classifiers experiments.
0. Setup
Before compiling Platform.
Miniconda
To be able to run Python Classifiers such as STree, ODTE, SVC, etc. it is needed to install Miniconda. To do so, download the installer from Miniconda and run it. It is recommended to install it in the home folder.
In Linux sometimes the library libstdc++ is mistaken from the miniconda installation and produces the next message when running the b_xxxx executables:
libstdc++.so.6: version `GLIBCXX_3.4.32' not found (required by b_xxxx)
The solution is to erase the libstdc++ library from the miniconda installation and no further compilation is needed.
MPI
In Linux just install openmpi & openmpi-devel packages.
source /etc/profile.d/modules.sh
module load mpi/openmpi-x86_64
If cmake can't find openmpi installation (like in Oracle Linux) set the following variable:
export MPI_HOME="/usr/lib64/openmpi"
In Mac OS X, install mpich with brew and if cmake doesn't find it, edit mpicxx wrapper to remove the ",-commons,use_dylibs" from final_ldflags
vi /opt/homebrew/bin/mpicxx
boost library
The best option is install the packages that the Linux distribution have in its repository. If this is the case:
sudo dnf install boost-devel
If this is not possible and the compressed packaged is installed, the following environment variable has to be set pointing to the folder where it was unzipped to:
export BOOST_ROOT=/path/to/library/
In some cases, it is needed to build the library, to do so:
cd /path/to/library
mkdir own
./bootstrap.sh --prefix=/path/to/library/own
./b2 install
export BOOST_ROOT=/path/to/library/own/
Don't forget to add the export BOOST_ROOT statement to .bashrc or wherever it is meant to be.
libxlswriter
cd lib/libxlsxwriter
make
make install DESTDIR=/home/rmontanana/Code PREFIX=
Release
make release
Debug & Tests
make debug
Configuration
The configuration file is named .env and it should be located in the folder where the experiments should be run. In the root folder of the project there is a file named .env.example that can be used as a template.
1. Commands
b_list
List all the datasets and its properties. The datasets are located in the datasets folder under the experiments root folder. A special file called all.txt with the names of the datasets has to be created. This all file is built wih lines of the form: ,<class_name>,<real_features>
where <real_features> can be either the word all or a list of numbers separated by commas, i.e. [0,3,6,7]
b_grid
Run a grid search over the parameters of the classifiers. The parameters are defined in the file grid.txt located in the grid folder of the experiments. The file has to be created with the following format:
{
"all": [
<set of hyperparams>, ...
],
"<dataset_name>": [
<specific set of hyperparams for <dataset_name>>, ...
],
}
The file has to be named grid<model_name>input.json
As a result it builds a file named grid<model_name>output.json with the results of the grid search.
The computation is done in parallel using MPI.
b_main
Run the main experiment. There are several hyperparameters that can set in command line:
- -d, --dataset <dataset_name> : Name of the dataset to run the experiment with. If no dataset is specificied the experiment will run with all the datasets in the all.txt file.
- -m, --model <classifier_name> : Name of the classifier to run the experiment with (i.e. BoostAODE, TAN, Odte, etc.).
- --discretize: Discretize the dataset before running the experiment.
- --stratified: Use stratified cross validation.
- --folds : Number of folds for cross validation (optional, default value is in .env file).
- -s, --seeds : Seeds for the random number generator (optional, default values are in .env file).
- --no-train-score: Do not calculate the train score (optional), this is useful when the dataset is big and the training score is not needed.
- --hyperparameters : Hyperparameters for the experiment in json format.
- --hyper-file <hyperparameters_file>: File with the hyperparameters for the experiment in json format. This file uses the output format of the b_grid command.
- --title <title_text>: Title of the experiment (optional if only one dataset is specificied).
- --quiet: Don't display detailed progress and result of the experiment.
b_manage
Manage the results of the experiments.
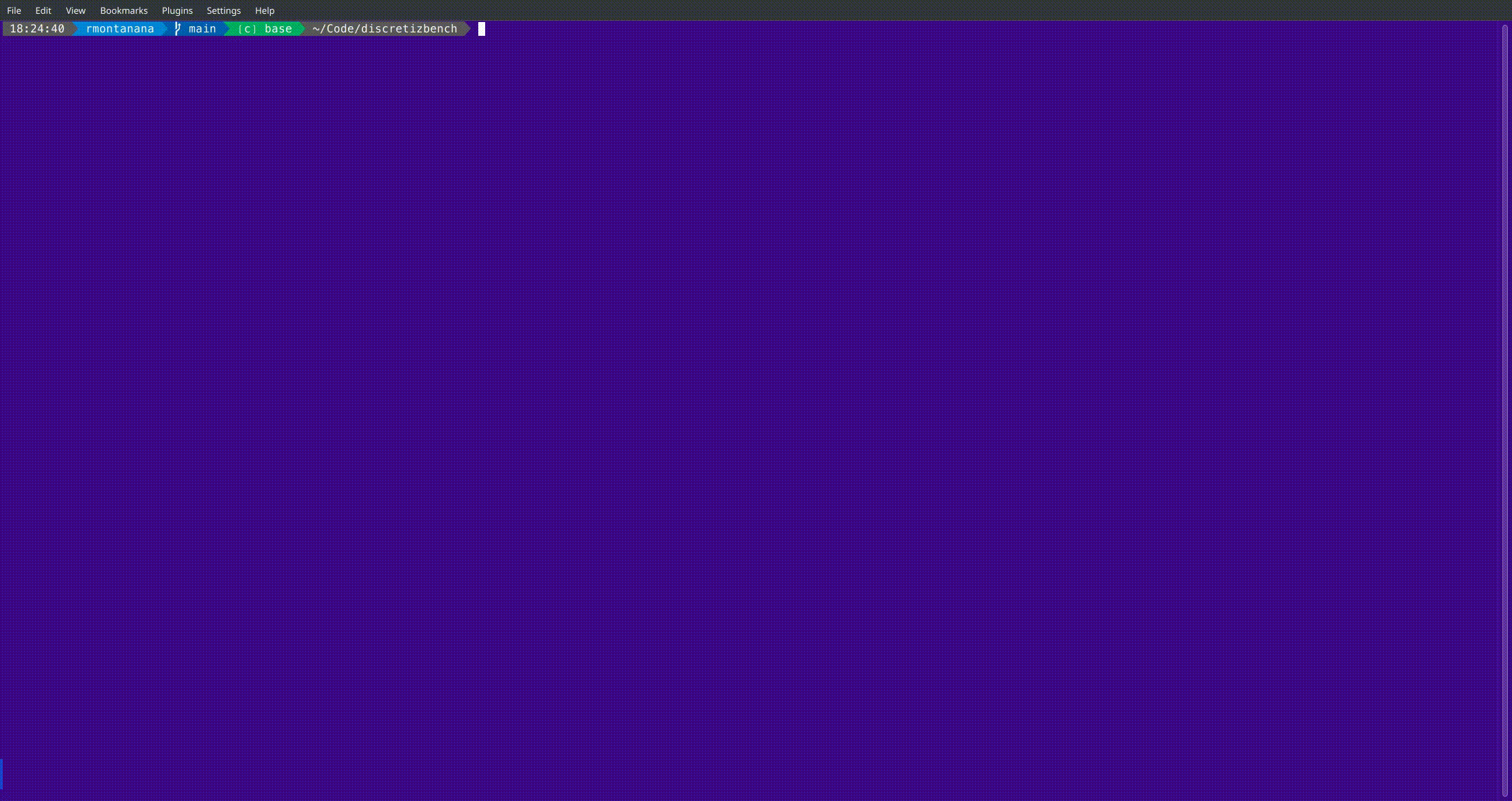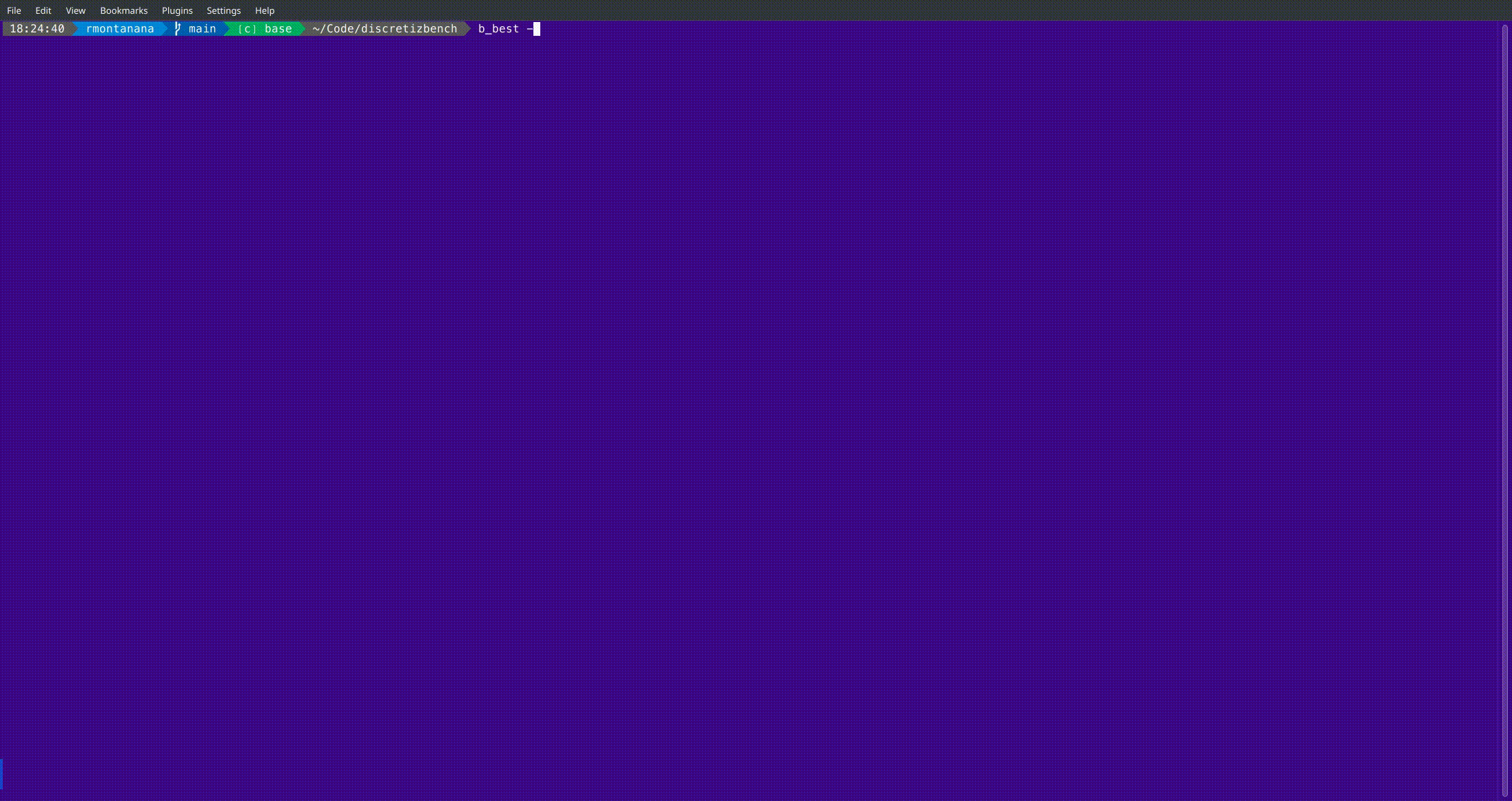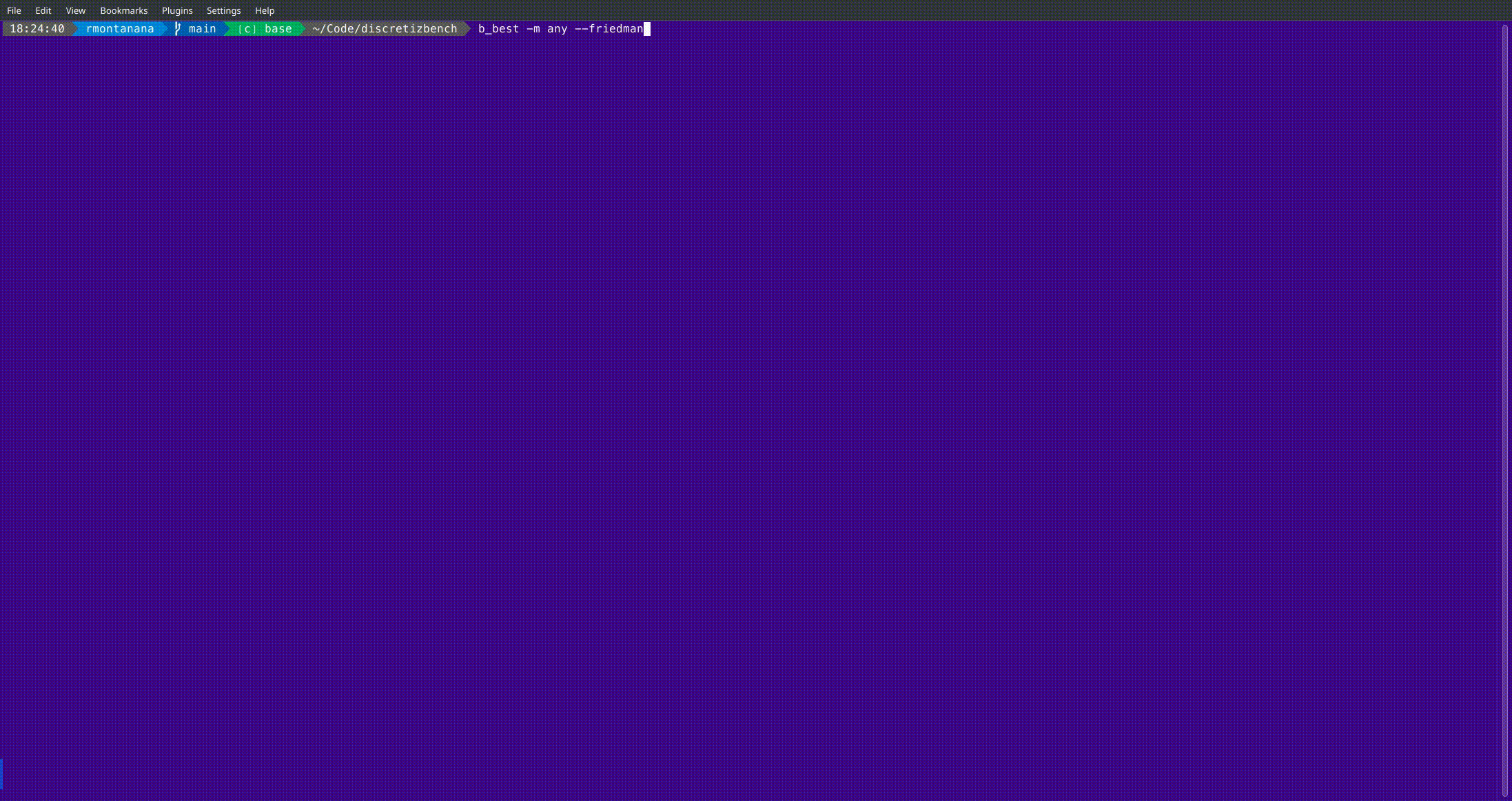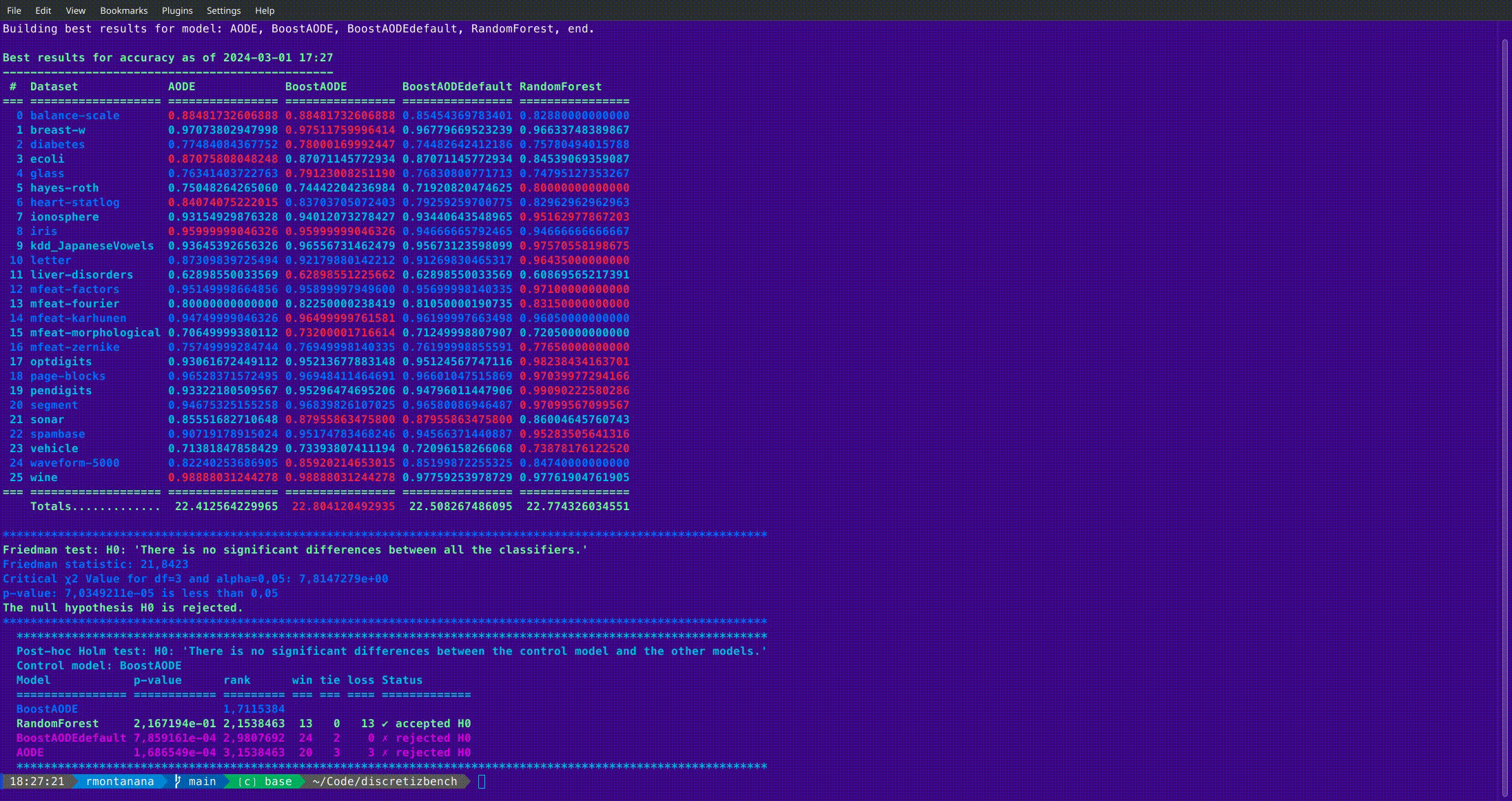
b_best
Get and optionally compare the best results of the experiments. The results can be stored in an MS Excel file.