#  Platform

[]()

Platform to run Bayesian Networks and Machine Learning Classifiers experiments.
## 0. Setup
Before compiling Platform.
### Miniconda
To be able to run Python Classifiers such as STree, ODTE, SVC, etc. it is needed to install Miniconda. To do so, download the installer from [Miniconda](https://docs.conda.io/en/latest/miniconda.html) and run it. It is recommended to install it in the home folder.
In Linux sometimes the library libstdc++ is mistaken from the miniconda installation and produces the next message when running the b_xxxx executables:
```bash
libstdc++.so.6: version `GLIBCXX_3.4.32' not found (required by b_xxxx)
```
The solution is to erase the libstdc++ library from the miniconda installation:
### MPI
In Linux just install openmpi & openmpi-devel packages. Only if cmake can't find openmpi installation (like in Oracle Linux) set the following variable:
```bash
export MPI_HOME="/usr/lib64/openmpi"
```
In Mac OS X, install mpich with brew and if cmake doesn't find it, edit mpicxx wrapper to remove the ",-commons,use_dylibs" from final_ldflags
```bash
vi /opt/homebrew/bin/mpicx
```
### boost library
[Getting Started]()
The best option is install the packages that the Linux distribution have in its repository. If this is the case:
```bash
sudo dnf install boost-devel
```
If this is not possible and the compressed packaged is installed, the following environment variable has to be set pointing to the folder where it was unzipped to:
```bash
export BOOST_ROOT=/path/to/library/
```
In some cases, it is needed to build the library, to do so:
```bash
cd /path/to/library
mkdir own
./bootstrap.sh --prefix=/path/to/library/own
./b2 install
export BOOST_ROOT=/path/to/library/own/
```
Don't forget to add the export BOOST_ROOT statement to .bashrc or wherever it is meant to be.
### libxlswriter
```bash
cd lib/libxlsxwriter
make
make install DESTDIR=/home/rmontanana/Code PREFIX=
```
### Release
```bash
make release
```
### Debug & Tests
```bash
make debug
```
### Configuration
The configuration file is named .env and it should be located in the folder where the experiments should be run. In the root folder of the project there is a file named .env.example that can be used as a template.
## 1. Commands
### b_list
List all the datasets and its properties. The datasets are located in the _datasets_ folder under the experiments root folder. A special file called all.txt with the names of the datasets has to be created. This all file is built wih lines of the form:
,,
where can be either the word _all_ or a list of numbers separated by commas, i.e. [0,3,6,7]
### b_grid
Run a grid search over the parameters of the classifiers. The parameters are defined in the file _grid.txt_ located in the grid folder of the experiments. The file has to be created with the following format:
```json
{
"all": [
, ...
],
"": [
>, ...
],
}
```
The file has to be named _grid__input.json_
As a result it builds a file named _grid__output.json_ with the results of the grid search.
The computation is done in parallel using MPI.

### b_main
Run the main experiment. There are several hyperparameters that can set in command line:
- -d, -\-dataset : Name of the dataset to run the experiment with. If no dataset is specificied the experiment will run with all the datasets in the all.txt file.
- -m, -\-model : Name of the classifier to run the experiment with (i.e. BoostAODE, TAN, Odte, etc.).
- -\-discretize: Discretize the dataset before running the experiment.
- -\-stratified: Use stratified cross validation.
- -\-folds : Number of folds for cross validation (optional, default value is in .env file).
- -s, -\-seeds : Seeds for the random number generator (optional, default values are in .env file).
- -\-no-train-score: Do not calculate the train score (optional), this is useful when the dataset is big and the training score is not needed.
- -\-hyperparameters : Hyperparameters for the experiment in json format.
- -\-hyper-file : File with the hyperparameters for the experiment in json format. This file uses the output format of the b_grid command.
- -\-title : Title of the experiment (optional if only one dataset is specificied).
- -\-quiet: Don't display detailed progress and result of the experiment.
### b_manage
Manage the results of the experiments.
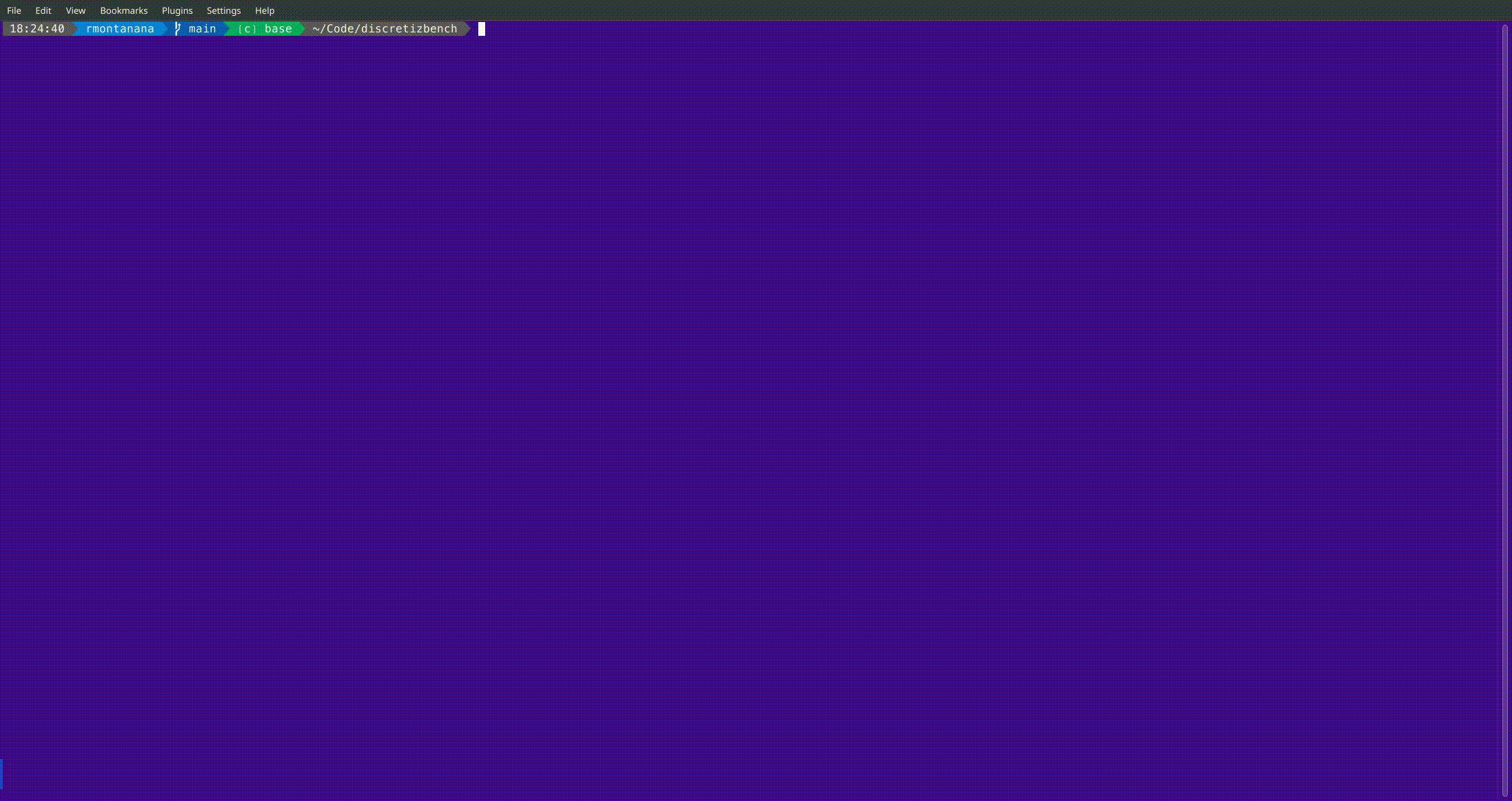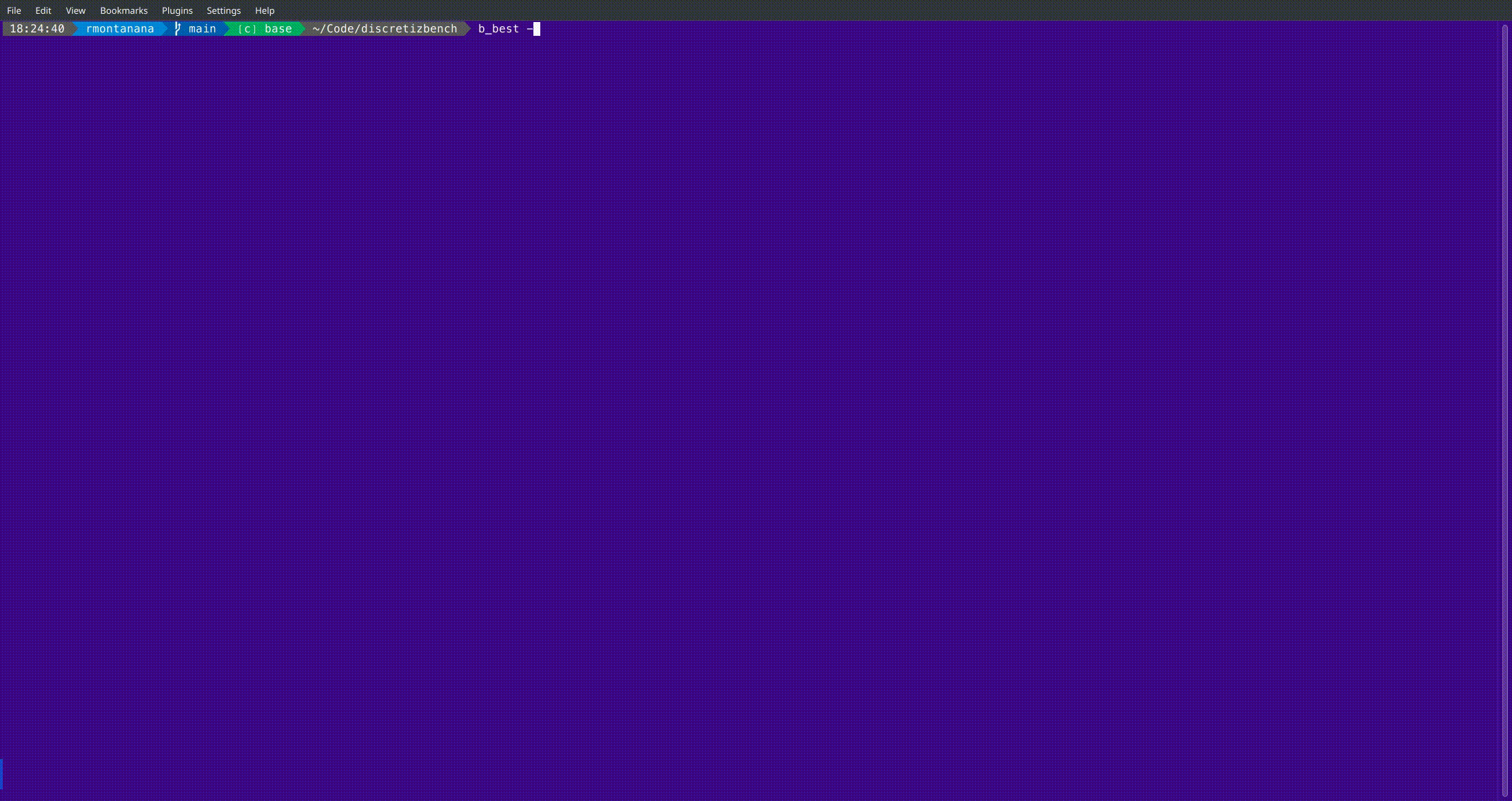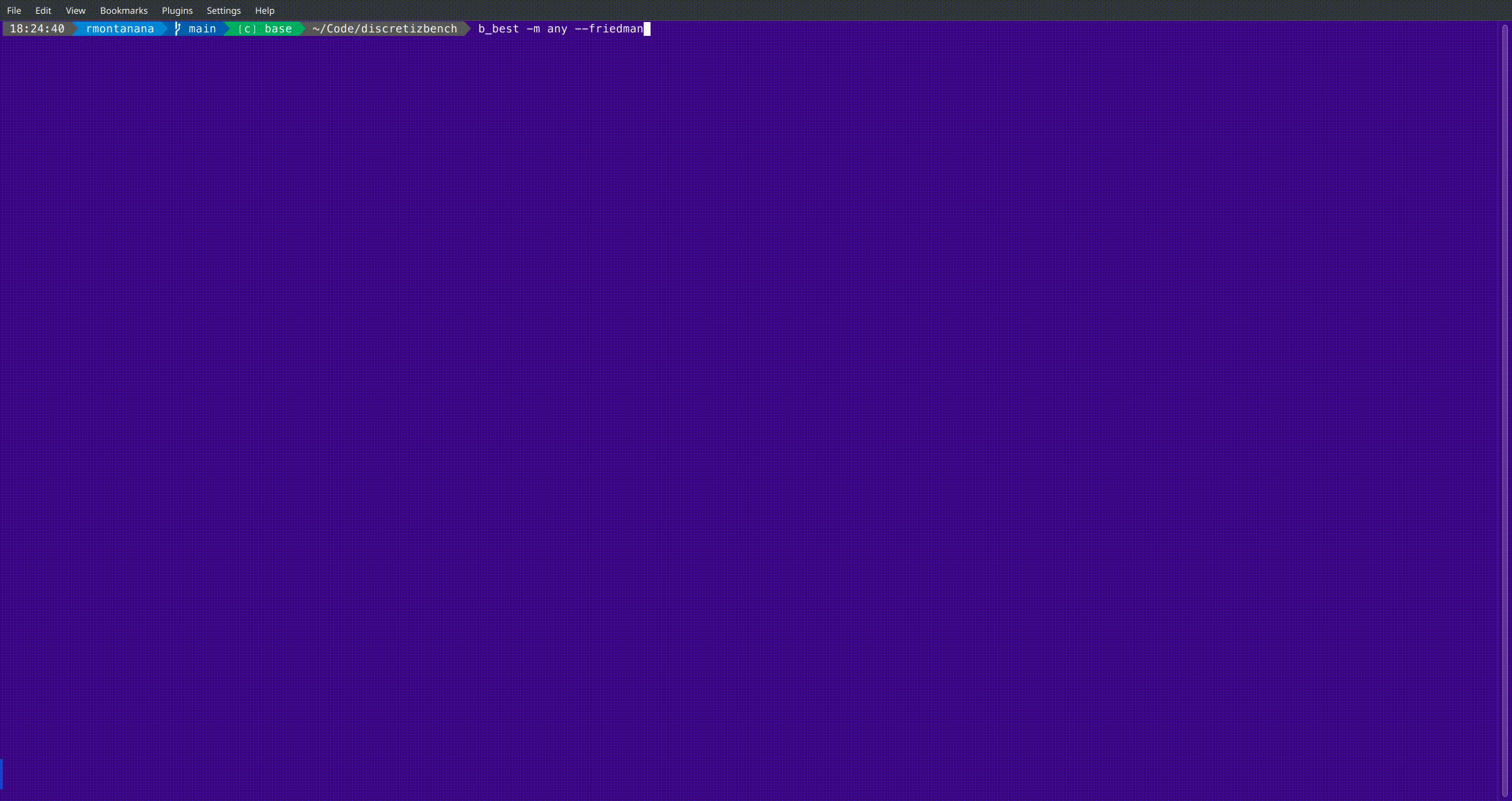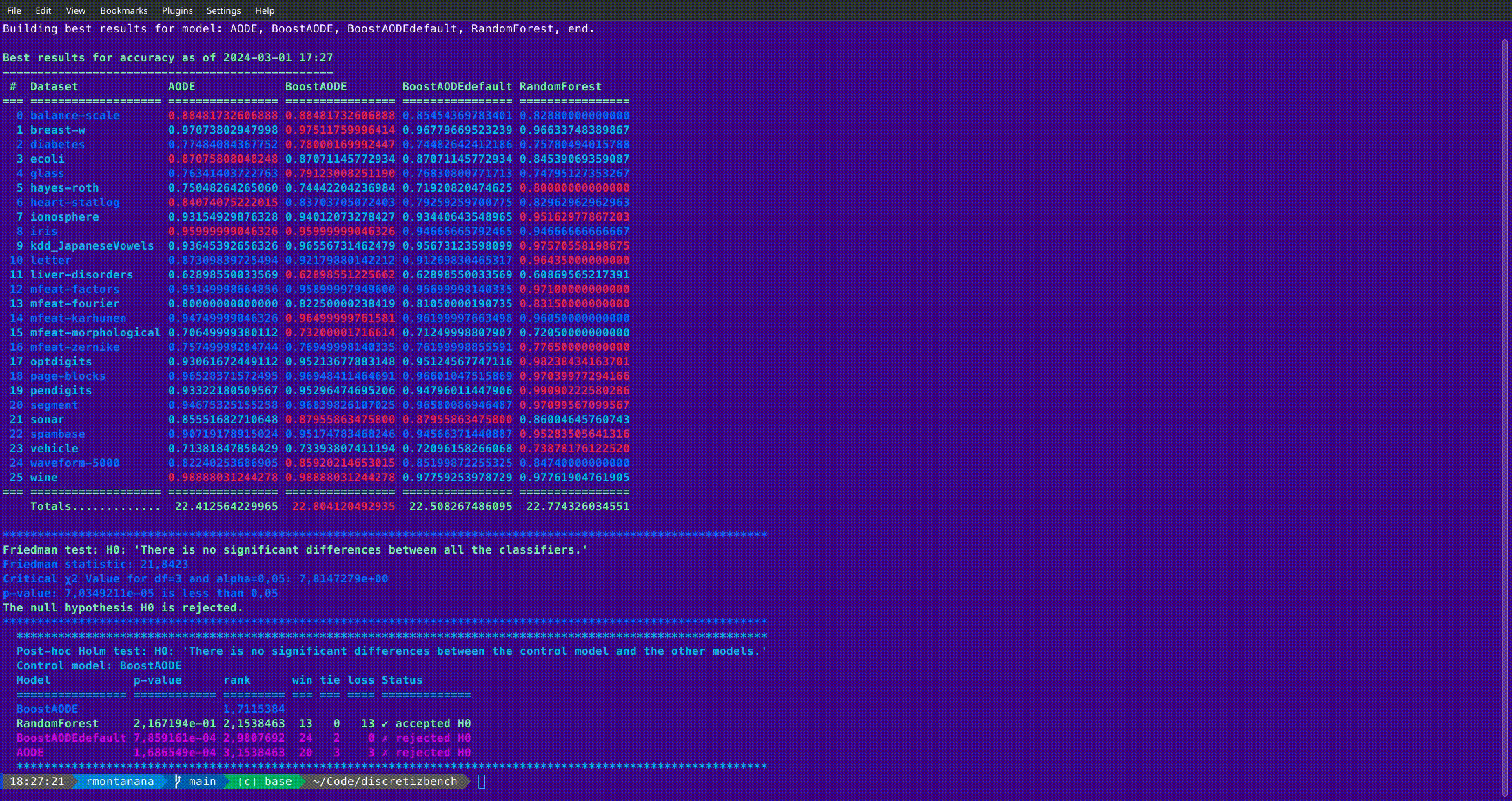
### b_best
Get and optionally compare the best results of the experiments. The results can be stored in an MS Excel file.
Platform

[]()

Platform to run Bayesian Networks and Machine Learning Classifiers experiments.
## 0. Setup
Before compiling Platform.
### Miniconda
To be able to run Python Classifiers such as STree, ODTE, SVC, etc. it is needed to install Miniconda. To do so, download the installer from [Miniconda](https://docs.conda.io/en/latest/miniconda.html) and run it. It is recommended to install it in the home folder.
In Linux sometimes the library libstdc++ is mistaken from the miniconda installation and produces the next message when running the b_xxxx executables:
```bash
libstdc++.so.6: version `GLIBCXX_3.4.32' not found (required by b_xxxx)
```
The solution is to erase the libstdc++ library from the miniconda installation:
### MPI
In Linux just install openmpi & openmpi-devel packages. Only if cmake can't find openmpi installation (like in Oracle Linux) set the following variable:
```bash
export MPI_HOME="/usr/lib64/openmpi"
```
In Mac OS X, install mpich with brew and if cmake doesn't find it, edit mpicxx wrapper to remove the ",-commons,use_dylibs" from final_ldflags
```bash
vi /opt/homebrew/bin/mpicx
```
### boost library
[Getting Started]()
The best option is install the packages that the Linux distribution have in its repository. If this is the case:
```bash
sudo dnf install boost-devel
```
If this is not possible and the compressed packaged is installed, the following environment variable has to be set pointing to the folder where it was unzipped to:
```bash
export BOOST_ROOT=/path/to/library/
```
In some cases, it is needed to build the library, to do so:
```bash
cd /path/to/library
mkdir own
./bootstrap.sh --prefix=/path/to/library/own
./b2 install
export BOOST_ROOT=/path/to/library/own/
```
Don't forget to add the export BOOST_ROOT statement to .bashrc or wherever it is meant to be.
### libxlswriter
```bash
cd lib/libxlsxwriter
make
make install DESTDIR=/home/rmontanana/Code PREFIX=
```
### Release
```bash
make release
```
### Debug & Tests
```bash
make debug
```
### Configuration
The configuration file is named .env and it should be located in the folder where the experiments should be run. In the root folder of the project there is a file named .env.example that can be used as a template.
## 1. Commands
### b_list
List all the datasets and its properties. The datasets are located in the _datasets_ folder under the experiments root folder. A special file called all.txt with the names of the datasets has to be created. This all file is built wih lines of the form:
,,
where can be either the word _all_ or a list of numbers separated by commas, i.e. [0,3,6,7]
### b_grid
Run a grid search over the parameters of the classifiers. The parameters are defined in the file _grid.txt_ located in the grid folder of the experiments. The file has to be created with the following format:
```json
{
"all": [
, ...
],
"": [
>, ...
],
}
```
The file has to be named _grid__input.json_
As a result it builds a file named _grid__output.json_ with the results of the grid search.
The computation is done in parallel using MPI.

### b_main
Run the main experiment. There are several hyperparameters that can set in command line:
- -d, -\-dataset : Name of the dataset to run the experiment with. If no dataset is specificied the experiment will run with all the datasets in the all.txt file.
- -m, -\-model : Name of the classifier to run the experiment with (i.e. BoostAODE, TAN, Odte, etc.).
- -\-discretize: Discretize the dataset before running the experiment.
- -\-stratified: Use stratified cross validation.
- -\-folds : Number of folds for cross validation (optional, default value is in .env file).
- -s, -\-seeds : Seeds for the random number generator (optional, default values are in .env file).
- -\-no-train-score: Do not calculate the train score (optional), this is useful when the dataset is big and the training score is not needed.
- -\-hyperparameters : Hyperparameters for the experiment in json format.
- -\-hyper-file : File with the hyperparameters for the experiment in json format. This file uses the output format of the b_grid command.
- -\-title : Title of the experiment (optional if only one dataset is specificied).
- -\-quiet: Don't display detailed progress and result of the experiment.
### b_manage
Manage the results of the experiments.
### b_best
Get and optionally compare the best results of the experiments. The results can be stored in an MS Excel file.

 Platform

[](
Platform

[](